Difference between revisions of "Category:Pathway"
From metabolic_network
(Created page with "* navigation ** mainpage|mainpage-description ** randompage-url|randompage ** Special:ListFiles|Files * Metabolic network components ** Category:Reaction|Reaction ** Category:...") |
(Created page with "== EFAEV583GEM description == == Automatic reconstruction with [http://aureme.genouest.org AuReMe] == Model summary: summary Download '''AuReMe''' Input...") |
||
| Line 1: | Line 1: | ||
| − | + | == EFAEV583GEM description == | |
| − | + | == Automatic reconstruction with [http://aureme.genouest.org AuReMe] == | |
| − | + | Model summary: [[MEDIA:summary.txt|summary]] | |
| − | + | ||
| − | + | Download '''AuReMe''' Input/Output [LINK OR MEDIA data] | |
| − | + | ||
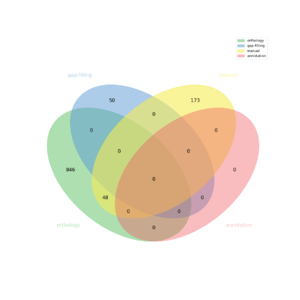
| − | * | + | The automatic reconstruction of ''Enterococcus_faecalis_str_v583'' results to a Genome scale [[MEDIA:model.xml|Model]] containing 1117 reactions, 911 metabolites, 610 genes and 106 pathways. This GeM was obtained based on the following sources: |
| − | ** | + | * Based on orthology data: |
| − | ** | + | ** Tool: [http://pathtastic.gforge.inria.fr Pantograph] |
| − | * | + | *** From template ''E_COLI_SSTR_K12_SUBSTR_MG1655'' creation of a metabolic network containing: 528 reactions |
| − | ** | + | *** From template ''L_PLANTARUM_WCFS1'' creation of a metabolic network containing: 385 reactions |
| − | ** | + | *** From template ''B_SUBTILIS_SUBSP_SUBTILIS_STR168'' creation of a metabolic network containing: 463 reactions |
| − | * | + | * Based on expertise: |
| − | * | + | *** 221 reaction(s) added |
| − | ** | + | * Based on gap-filling: |
| − | * | + | ** Tool: [https://pypi.python.org/pypi/meneco meneco] |
| − | * | + | *** 50 reaction(s) added |
| − | ** | + | |
| − | * | + | [[FILE:venn.png|frameless|border]] |
| − | ** added | + | |
| − | + | == Collaborative curation == | |
| − | + | * Suggest reactions to add or remove: | |
| − | + | ** Download this [[MEDIA:Add_delete_reaction.csv|form]] | |
| − | * | + | * Suggest new reactions to create and add: |
| − | ** | + | ** Download this [[MEDIA:Reaction_creator.csv|form]] |
| − | * | + | * '''Follow the examples given in the form(s) to correctly share your suggestions''' |
| − | ** | + | * Send the filled form(s) to: CONTACT_MAIL |
| − | * | + | |
Revision as of 18:51, 18 March 2018
EFAEV583GEM description
Automatic reconstruction with AuReMe
Model summary: summary
Download AuReMe Input/Output [LINK OR MEDIA data]
The automatic reconstruction of Enterococcus_faecalis_str_v583 results to a Genome scale Model containing 1117 reactions, 911 metabolites, 610 genes and 106 pathways. This GeM was obtained based on the following sources:
- Based on orthology data:
- Tool: Pantograph
- From template E_COLI_SSTR_K12_SUBSTR_MG1655 creation of a metabolic network containing: 528 reactions
- From template L_PLANTARUM_WCFS1 creation of a metabolic network containing: 385 reactions
- From template B_SUBTILIS_SUBSP_SUBTILIS_STR168 creation of a metabolic network containing: 463 reactions
- Tool: Pantograph
- Based on expertise:
- 221 reaction(s) added
- Based on gap-filling:
- Tool: meneco
- 50 reaction(s) added
- Tool: meneco
Collaborative curation
- Suggest reactions to add or remove:
- Download this form
- Suggest new reactions to create and add:
- Download this form
- Follow the examples given in the form(s) to correctly share your suggestions
- Send the filled form(s) to: CONTACT_MAIL
Pages in category "Pathway"
The following 106 pages are in this category, out of 106 total.
M
- Map00010
- Map00020
- Map00030
- Map00040
- Map00051
- Map00052
- Map00053
- Map00061
- Map00071
- Map00072
- Map00130
- Map00190
- Map00220
- Map00230
- Map00240
- Map00250
- Map00254
- Map00340
- Map00261
- Map00270
- Map00280
- Map00290
- Map01051
- Map00300
- Map00310
- Map00330
- Map00332
- Map00350
- Map00360
- Map00362
- Map00380
- Map00400
- Map00401
- Map00410
- Map00430
- Map00450
- Map00460
- Map00471
- Map00473
- Map00480
- Map00500
- Map00511
- Map00520
- Map00521
- Map00523
- Map00524
- Map00525
- Map00531
- Map00540
- Map00550
- Map00561
- Map00562
- Map00564
- Map00600
- Map00604
- Map00620
- Map00621
- Map00622
- Map00623
- Map00625
- Map00627
- Map00630
- Map00640
- Map00650
- Map00660
- Map00670
- Map00680
- Map00710
- Map00720
- Map00730
- Map00740
- Map00750
- Map00760
- Map00770
- Map00780
- Map00785
- Map00790
- Map00860
- Map00900
- Map00901
- Map00903
- Map00910
- Map00920
- Map00950
- Map00960
- Map00965
- Map00966
- Map00970
- Map00980
- Map00981
- Map00982
- Map01040
- Map00983
- Map01053
- Map01055
- Map01100
- Map01110
- Map01120
- Map01130
- Map01200
- Map01210
- Map01212
- Map01220
- Map01230
- Map04070