Difference between revisions of "Ec-11 002430"
From metabolic_network
(→Automatic reconstruction with AuReMe) |
|||
| Line 3: | Line 3: | ||
Model summary: [[MEDIA:ESiliculosus_final.txt|summary]] | Model summary: [[MEDIA:ESiliculosus_final.txt|summary]] | ||
| − | Download '''AuReMe''' Input/Output [https://mybox.inria.fr data] | + | Download '''AuReMe''' Input/Output [https://mybox.inria.fr/f/715c7378e9/?raw=1 data] |
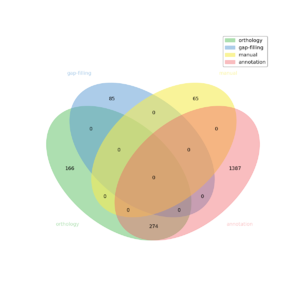
The automatic reconstruction of ''Ectocarpus_Siliculosus'' results to a Genome scale [[MEDIA:ESiliculosus_final_lvl3.xml|Model]] containing 1977 reactions, 2132 metabolites, 2281 genes and 1101 pathways. This GeM was obtained based on the following sources: | The automatic reconstruction of ''Ectocarpus_Siliculosus'' results to a Genome scale [[MEDIA:ESiliculosus_final_lvl3.xml|Model]] containing 1977 reactions, 2132 metabolites, 2281 genes and 1101 pathways. This GeM was obtained based on the following sources: | ||
Revision as of 14:26, 12 January 2018
ECTOGEM description
Automatic reconstruction with AuReMe
Model summary: summary
Download AuReMe Input/Output data
The automatic reconstruction of Ectocarpus_Siliculosus results to a Genome scale Model containing 1977 reactions, 2132 metabolites, 2281 genes and 1101 pathways. This GeM was obtained based on the following sources:
- Based on annotation data:
- Tool: Pathway tools
- Creation of a metabolic network containing 1661 reactions
- Tool: Pathway tools
- Based on orthology data:
- Tool: Pantograph
- From template ARAGEM creation of a metabolic network containing: 440 reactions
- Tool: Pantograph
- Based on expertise:
- 65 reaction(s) added
- Based on gap-filling:
- Tool: meneco
- 85 reaction(s) added
- Tool: meneco