Difference between revisions of "Gap-filling-gapfilling solution with meneco draft medium"
From metabolic_network
(→Automatic reconstruction with AuReMe) |
(→Collaborative curation) |
||
| Line 29: | Line 29: | ||
** Download this [[MEDIA:Reaction_creator.csv|form]] | ** Download this [[MEDIA:Reaction_creator.csv|form]] | ||
* '''Follow the examples given in the form(s) to correctly share your suggestions''' | * '''Follow the examples given in the form(s) to correctly share your suggestions''' | ||
| − | * Send the filled form(s) to: gem-aureme@ | + | * Send the filled form(s) to: gem-aureme@inria.fr |
Revision as of 14:24, 12 January 2018
TISOGEM description
Automatic reconstruction with AuReMe
Model summary: summary
Download AuReMe Input/Output data
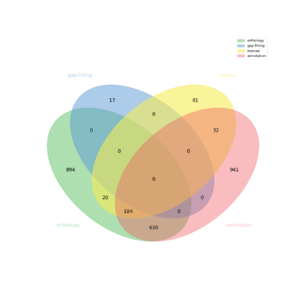
The automatic reconstruction of Tisocrysis_lutea results to a Genome scale Model containing 2796 reactions, 2745 metabolites, 2728 genes and 1212 pathways. This GeM was obtained based on the following sources:
- Based on annotation data:
- Tool: Pathway tools
- Creation of a metabolic network containing 2568 reactions
- Tool: Pathway tools
- Based on orthology data:
- Tool: Pantograph
- From template CREINHARDTII creation of a metabolic network containing: 558 reactions
- From template SYNECHOCYSTIS creation of a metabolic network containing: 260 reactions
- From template ATHALIANA creation of a metabolic network containing: 419 reactions
- From template ESILICULOSUS creation of a metabolic network containing: 1191 reactions
- Tool: Pantograph
- Based on expertise:
- 314 reaction(s) added
- Based on gap-filling:
- Tool: meneco
- 17 reaction(s) added
- Tool: meneco