Difference between revisions of "Main Page"
From metabolic_network
(Created page with "Category:Reaction == Reaction [http://metacyc.org/META/NEW-IMAGE?object=6PGLUCONDEHYDROG-RXN 6PGLUCONDEHYDROG-RXN] == * direction: ** LEFT-TO-RIGHT * common name: ** phosp...") |
|||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | == TISOGEM description == | |
| − | == | + | == Automatic reconstruction with [http://aureme.genouest.org AuReMe] == |
| − | + | Model summary: [[MEDIA:summary.txt|summary]] | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | Download '''AuReMe''' Input/Output [LINK OR MEDIA data] | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | The automatic reconstruction of ''Tisocrysis_lutea'' results to a Genome scale [[MEDIA:model.xml|Model]] containing 2799 reactions, 2747 metabolites, 2728 genes and 1212 pathways. This GeM was obtained based on the following sources: | |
| − | + | * Based on annotation data: | |
| − | + | ** Tool: [http://bioinformatics.ai.sri.com/ptools/ Pathway tools] | |
| − | + | *** Creation of a metabolic network containing 2568 reactions | |
| − | + | * Based on orthology data: | |
| − | + | ** Tool: [http://pathtastic.gforge.inria.fr Pantograph] | |
| − | + | *** From template ''CREINHARDTII'' creation of a metabolic network containing: 558 reactions | |
| − | + | *** From template ''SYNECHOCYSTIS'' creation of a metabolic network containing: 260 reactions | |
| − | + | *** From template ''ATHALIANA'' creation of a metabolic network containing: 419 reactions | |
| − | + | *** From template ''ESILICULOSUS'' creation of a metabolic network containing: 1191 reactions | |
| − | + | * Based on expertise: | |
| − | + | *** 317 reaction(s) added | |
| − | + | * Based on gap-filling: | |
| − | + | ** Tool: [https://pypi.python.org/pypi/meneco meneco] | |
| − | + | *** 17 reaction(s) added | |
| − | + | ||
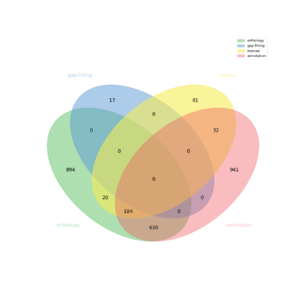
| − | + | [[FILE:venn.png|frameless|border]] | |
| − | * | + | |
| − | ** | + | == Collaborative curation == |
| − | + | * Suggest reactions to add or remove: | |
| − | + | ** Download this [[MEDIA:Add_delete_reaction.csv|form]] | |
| − | + | * Suggest new reactions to create and add: | |
| − | + | ** Download this [[MEDIA:Reaction_creator.csv|form]] | |
| − | + | * '''Follow the examples given in the form(s) to correctly share your suggestions''' | |
| − | + | * Send the filled form(s) to: gem-aureme@inria.fr | |
| − | * | + | |
| − | ** | + | |
| − | * | + | |
| − | ** | + | |
| − | + | ||
| − | + | ||
| − | ** | + | |
| − | * | + | |
| − | ** | + | |
| − | * | + | |
| − | ** | + | |
| − | * | + | |
| − | ** | + | |
| − | * | + | |
| − | * | + | |
| − | ** | + | |
| − | * | + | |
| − | * | + | |
| − | ** | + | |
| − | + | ||
| − | ** | + | |
| − | * | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | * | + | |
| − | ** [ | + | |
| − | * | + | |
| − | ** [ | + | |
| − | * | + | |
| − | * | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 14:11, 16 November 2018
TISOGEM description
Automatic reconstruction with AuReMe
Model summary: summary
Download AuReMe Input/Output [LINK OR MEDIA data]
The automatic reconstruction of Tisocrysis_lutea results to a Genome scale Model containing 2799 reactions, 2747 metabolites, 2728 genes and 1212 pathways. This GeM was obtained based on the following sources:
- Based on annotation data:
- Tool: Pathway tools
- Creation of a metabolic network containing 2568 reactions
- Tool: Pathway tools
- Based on orthology data:
- Tool: Pantograph
- From template CREINHARDTII creation of a metabolic network containing: 558 reactions
- From template SYNECHOCYSTIS creation of a metabolic network containing: 260 reactions
- From template ATHALIANA creation of a metabolic network containing: 419 reactions
- From template ESILICULOSUS creation of a metabolic network containing: 1191 reactions
- Tool: Pantograph
- Based on expertise:
- 317 reaction(s) added
- Based on gap-filling:
- Tool: meneco
- 17 reaction(s) added
- Tool: meneco