Difference between revisions of "Apo-3-methylcrotonoyl-CoA-carbon-dioxide"
From metabolic_network
| Line 5: | Line 5: | ||
Download '''AuReMe''' Input/Output [LINK OR MEDIA data] | Download '''AuReMe''' Input/Output [LINK OR MEDIA data] | ||
| − | The automatic reconstruction of ''Chondrus crispus'' results to a Genome scale [[MEDIA:model.xml|Model]] containing | + | The automatic reconstruction of ''Chondrus crispus'' results to a Genome scale [[MEDIA:model.xml|Model]] containing 1943 reactions, 2053 metabolites, 1701 genes and 1035 pathways. This GeM was obtained based on the following sources: |
* Based on annotation data: | * Based on annotation data: | ||
** Tool: [http://bioinformatics.ai.sri.com/ptools/ Pathway tools] | ** Tool: [http://bioinformatics.ai.sri.com/ptools/ Pathway tools] | ||
Revision as of 13:10, 31 January 2018
GEM description
Automatic reconstruction with AuReMe
Model summary: summary
Download AuReMe Input/Output [LINK OR MEDIA data]
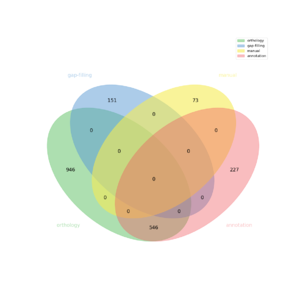
The automatic reconstruction of Chondrus crispus results to a Genome scale Model containing 1943 reactions, 2053 metabolites, 1701 genes and 1035 pathways. This GeM was obtained based on the following sources:
- Based on annotation data:
- Tool: Pathway tools
- Creation of a metabolic network containing 773 reactions
- Tool: Pathway tools
- Based on orthology data:
- Tool: Pantograph
- From template GALDIERIA.SULPHURARIA creation of a metabolic network containing: 1361 reactions
- From template A.TALIANA creation of a metabolic network containing: 383 reactions
- Tool: Pantograph
- Based on expertise:
- 73 reaction(s) added
- Based on gap-filling:
- Tool: meneco
- 151 reaction(s) added
- Tool: meneco
Collaborative curation
- Suggest reactions to add or remove:
- Download this form
- Suggest new reactions to create and add:
- Download this form
- Follow the examples given in the form(s) to correctly share your suggestions
- Send the filled form(s) to: gabriel.markov@sb-roscoff.fr
- For more information about this project, please contact Gabriel Markov (Station Biologique de Roscoff).